Step 1: Create a Project
Follow the steps below. Use ←/→ to navigate; press 1-9 to jump.
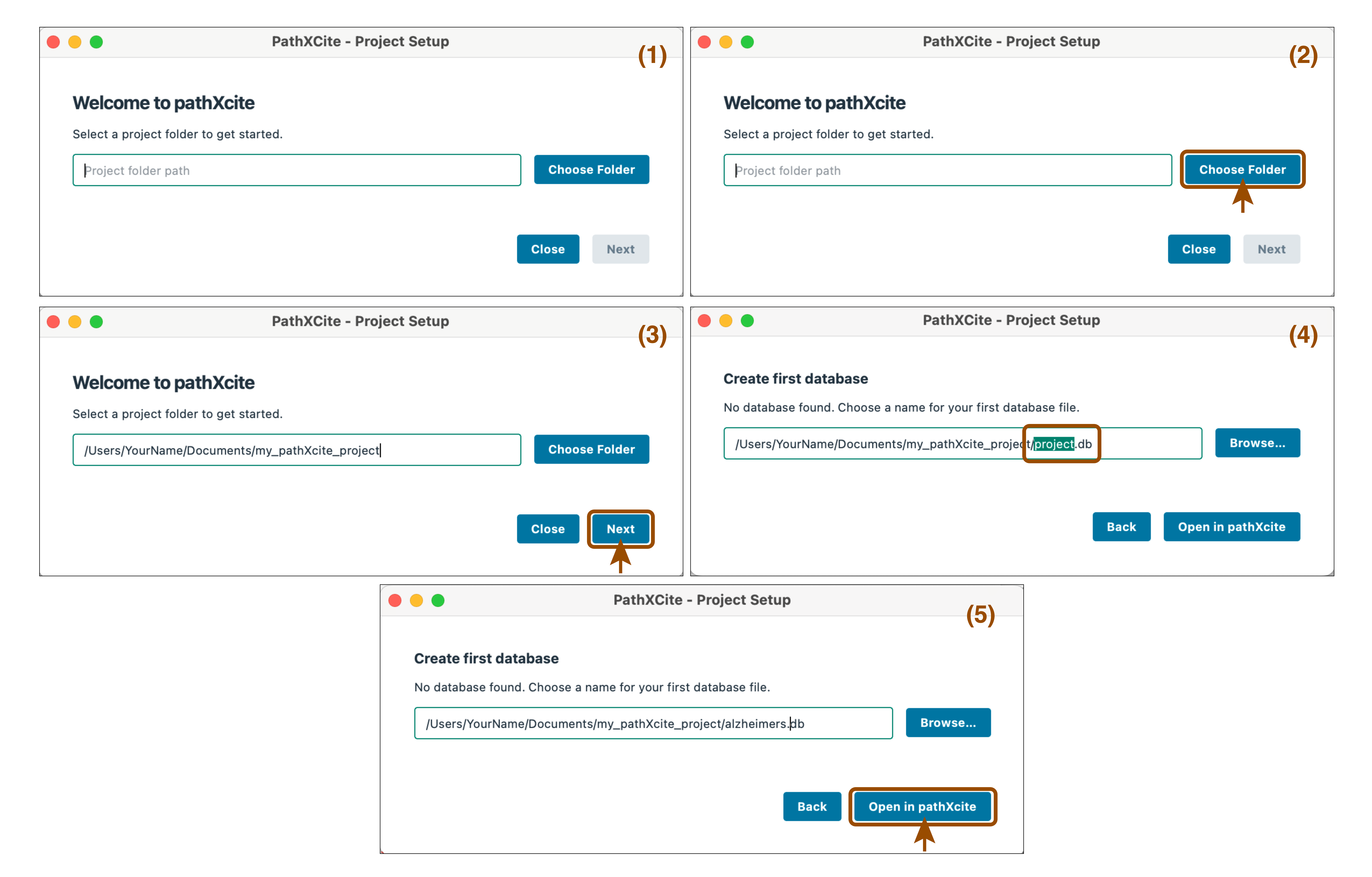
Launch the app and create/open a project so your local database, figures, and exports are saved together.
- Upon launching pathXcite, you will see the project setup screen.
- Select Choose Folder. Since we create a new project, choose an empty folder.
- Click Next.
- Now name your first database (e.g.,
alzheimers). - Click Open in pathXcite.
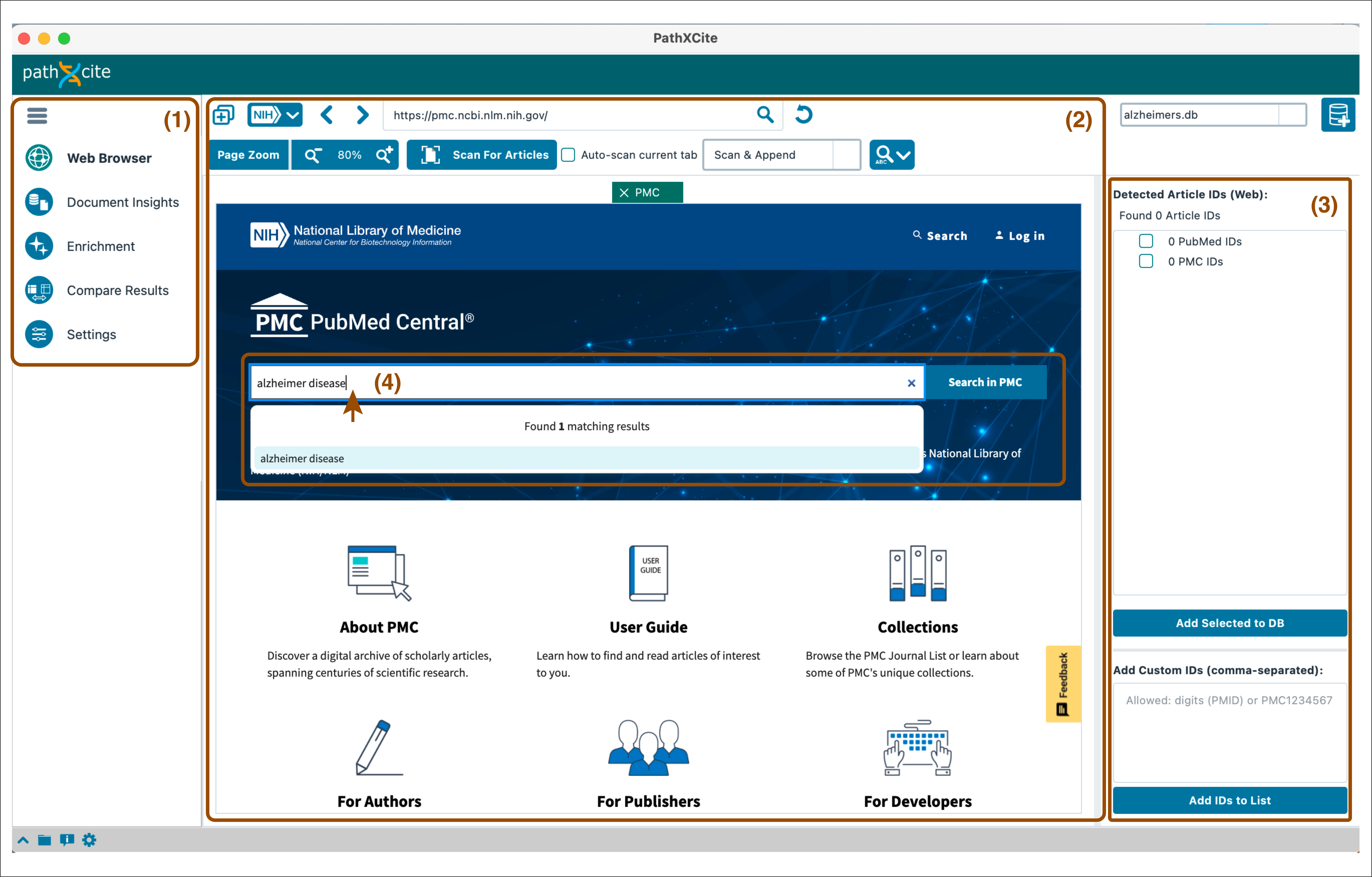
Query articles related to Alzheimer's disease.
- Now the main interface of pathXcite is visible. On the left you see the menu to switch between modules. We start with the Web Browser module (which should be already open after launching pathXcite).
- Open the Web Browser module, which holds an embedded browser in its main view.
- On the right, found article IDs will be listed. (Note: You can also add your own comma-separated list of IDs at the bottom of the list)
- Now, start your first query by searching
Alzheimer diseasein the search bar and press Enter.
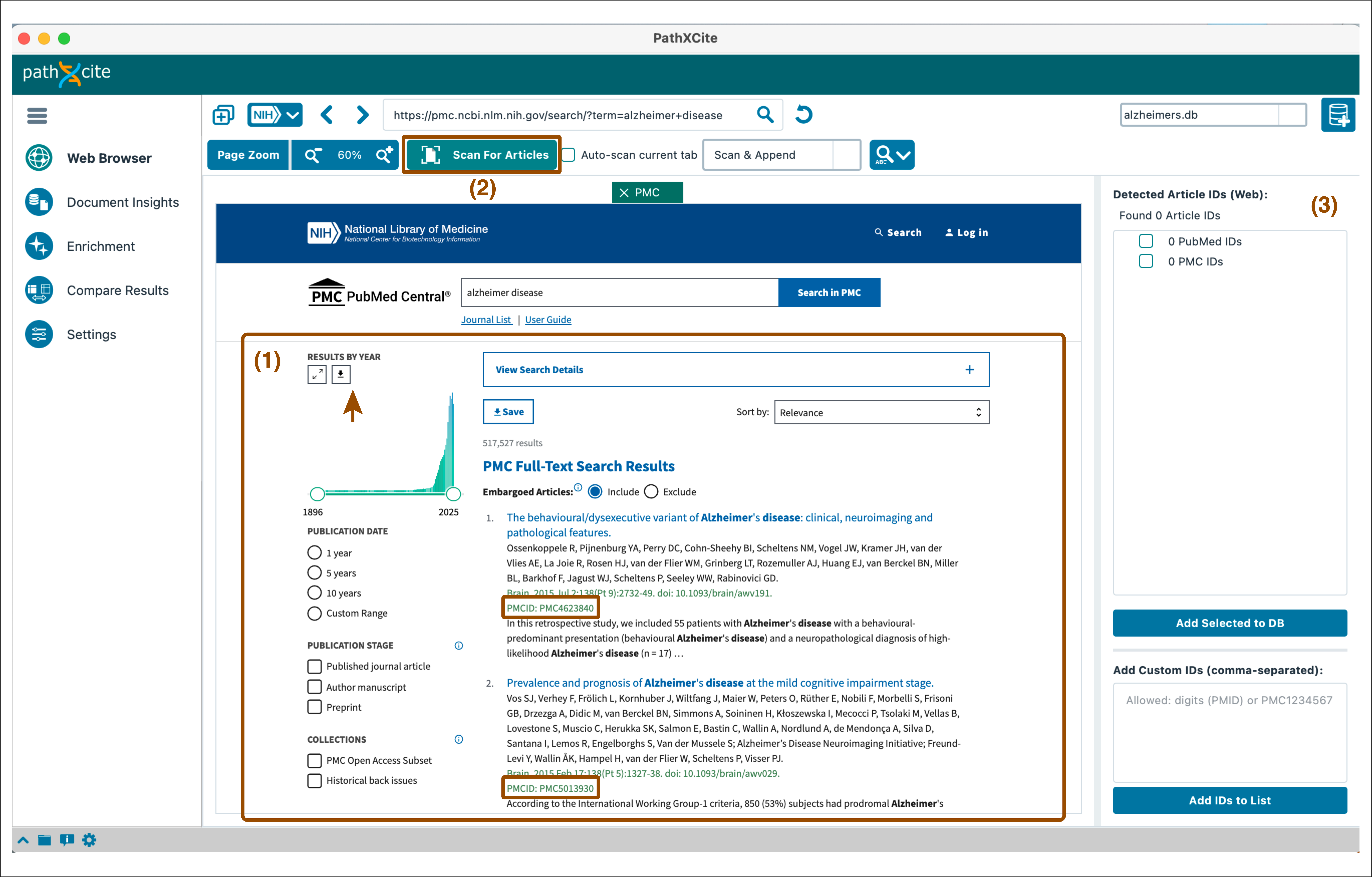
Scan the current page for article IDs.
- When using PubMed Central (or PubMed), the articles are listed with their respective IDs. On PubMed Central, initially the 10 most relevant articles are listed. Scroll down to select Show More Results to load additional articles.
- Clicking Scan For Articles automatically scans the current page for these IDs and will list them on the right.
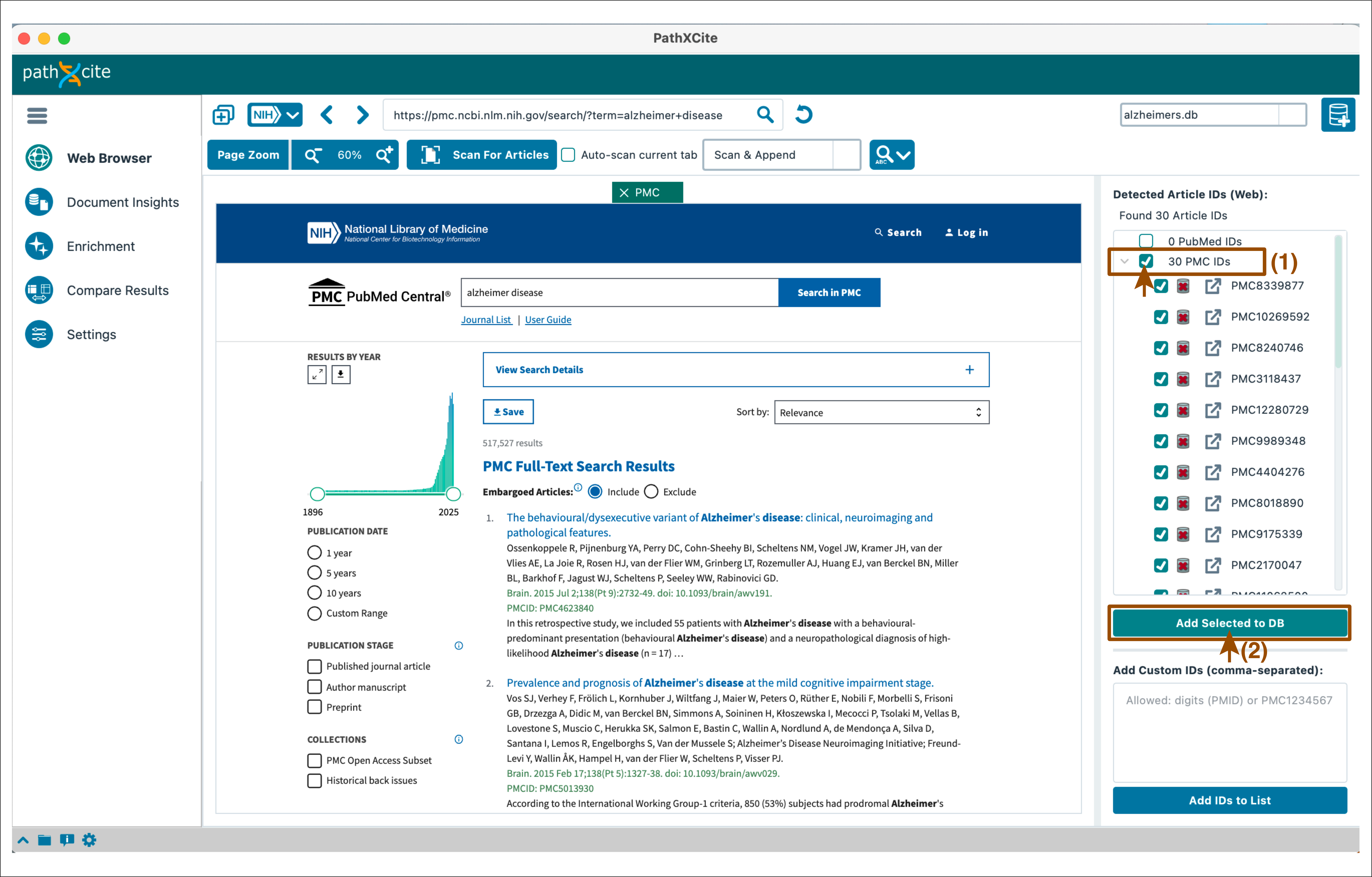
Retrieve the article metadata, genes mentioned and available text.
- In our example, the query result showed 30 PMC article IDs and no PubMed IDs in the web page. Select the articles you want to add to the current database. For now, we will select all.
- After selecting the articles, click Add Selected to DB. Depending on the number of selected articles and your network speed, this may take some seconds. For this small number of articles, it should not take longer than 20 seconds.
In the next step, we will get an overview of the articles we just added.
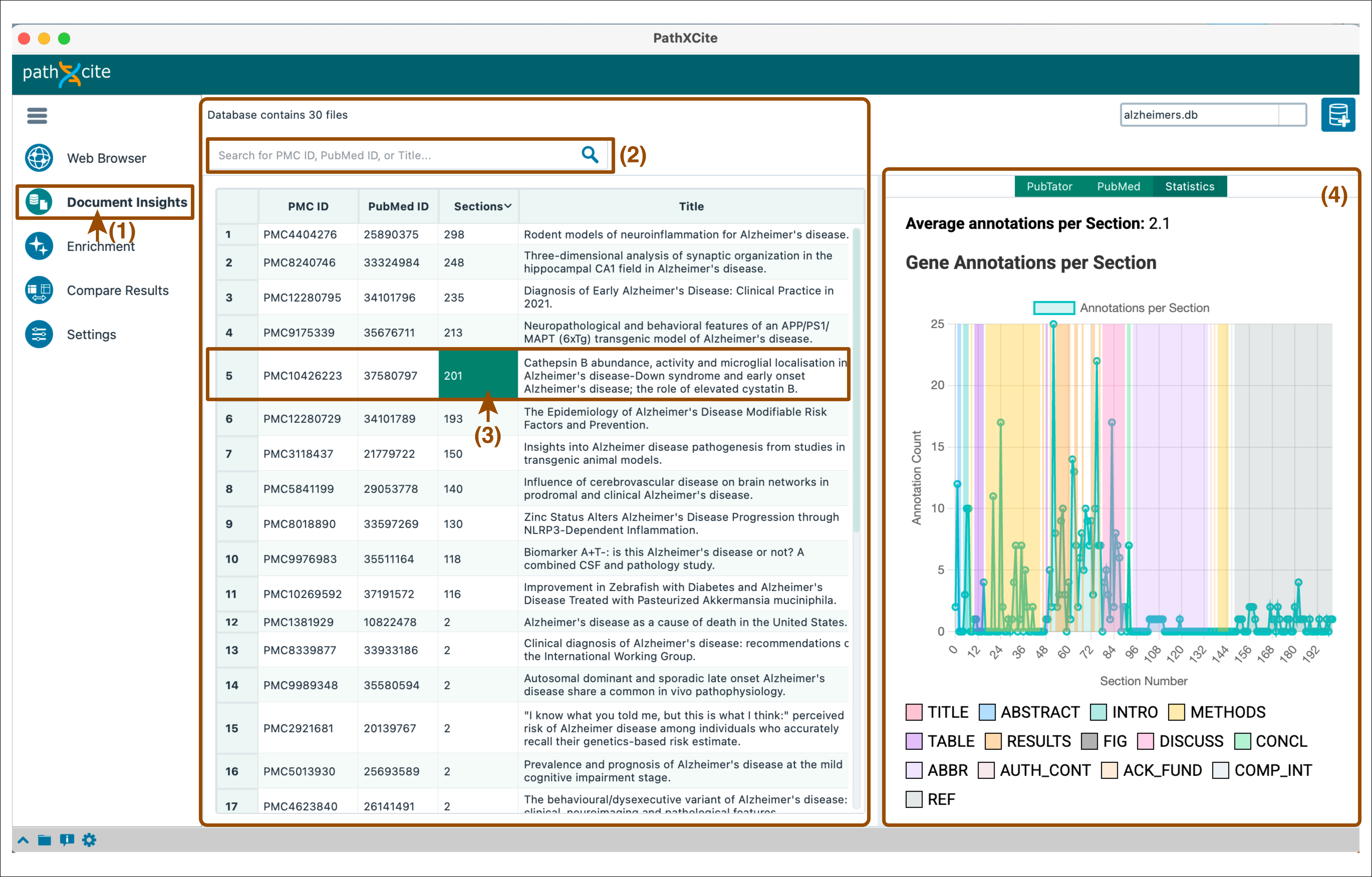
The Document Insights view provides an overview of the articles in your database, along with their metadata and annotations.
- Select Document Insights in the left menu.
- You can now see and explore all the articles that are in your current database, e.g. by searching their titles or specific IDs.
- Click on an article to see its metadata, available text, and gene annotations.
- On the right, you can find additional information and options for each article. You can on the one hand open the article within a small browser, or view the annotation statistics. The statistics plot shows the number of sections of the article on the x-axis, while the y-axis shows the number of genes found in each section. (See tutorial Analysing the Section Plot for more details.)
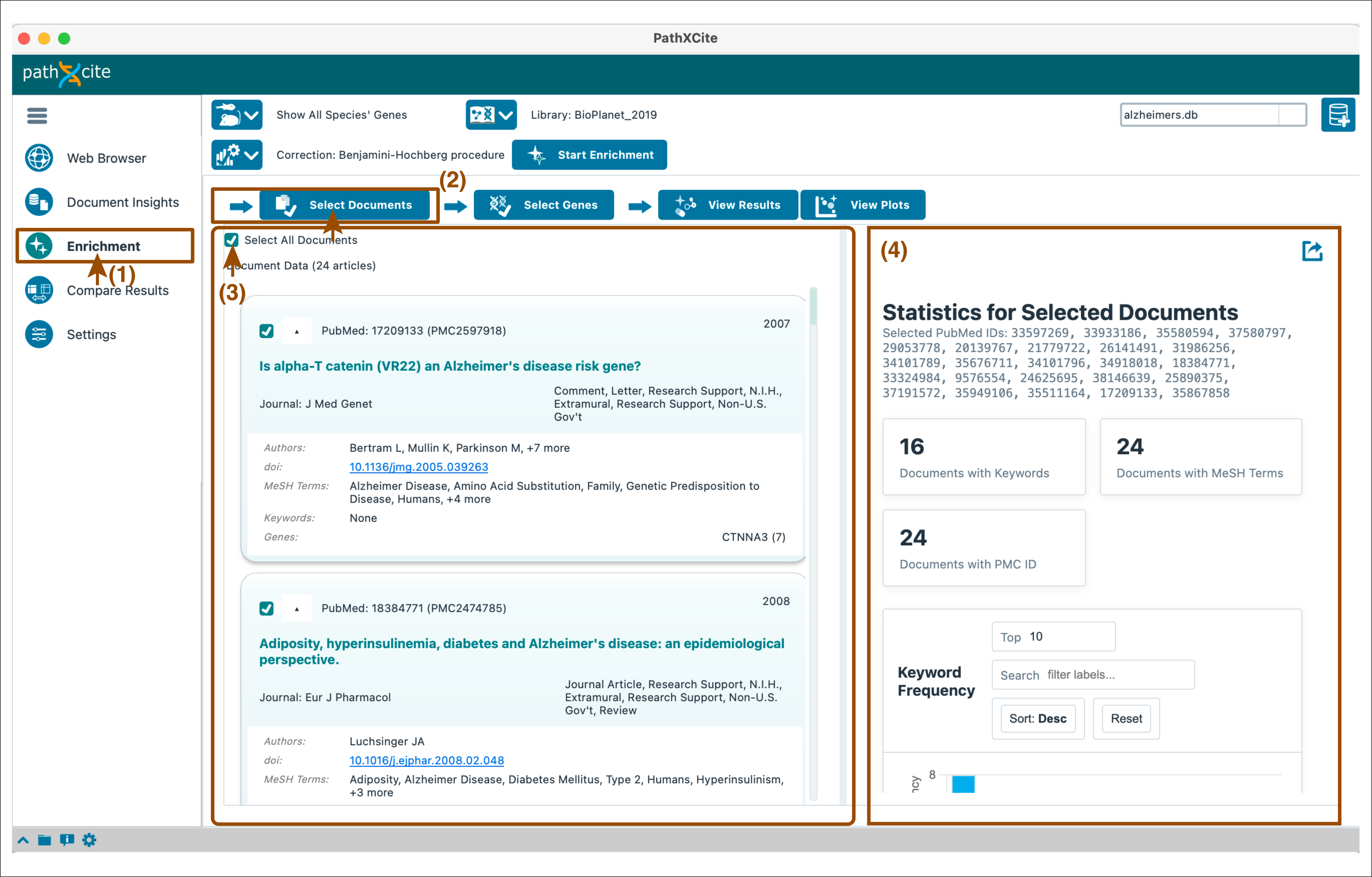
The first step of the enrichment analysis is to select the documents (articles).
Interactive document statistics are shown on the right. This can help you to identify relevant articles based on their content and context.
- Select Enrichment in the left menu.
- Click on Select Documents (this is the first step of the enrichment analysis; the arrows represent the pipeline we will follow).
- Select the documents (articles) you want to include. For now, we select all the documents. Note, that from our previous 30 articles, 24 mention at least one gene, while 6 were excluded.
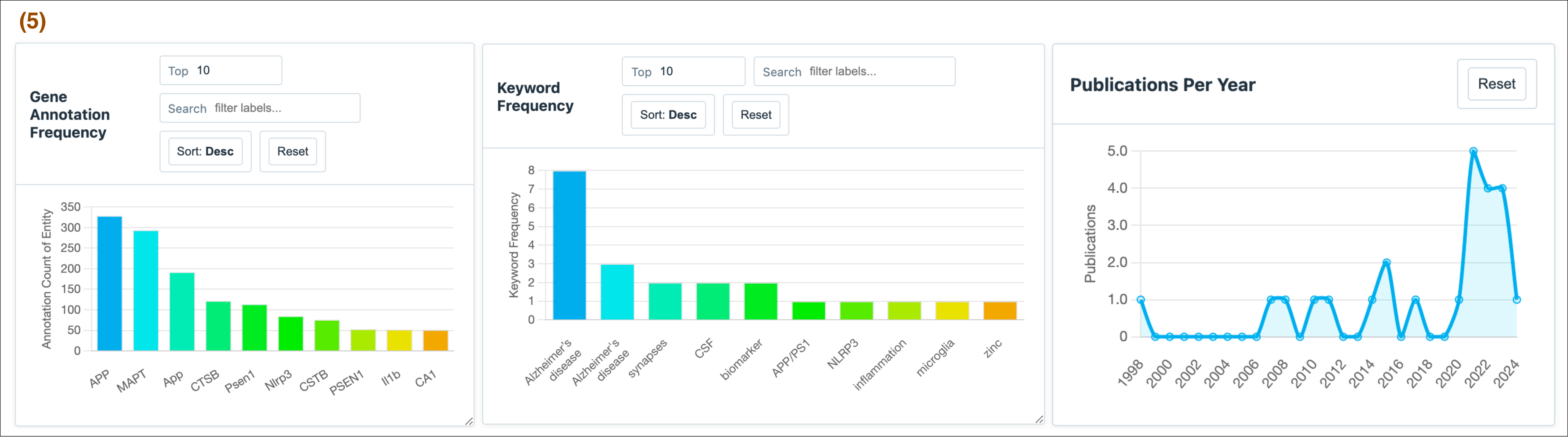
- Whenever you change the selection, the right view automatically updates the statistics. You can explore the most frequent keywords, MeSH terms, journals, genes, or investigate the distribution of articles per year (5).
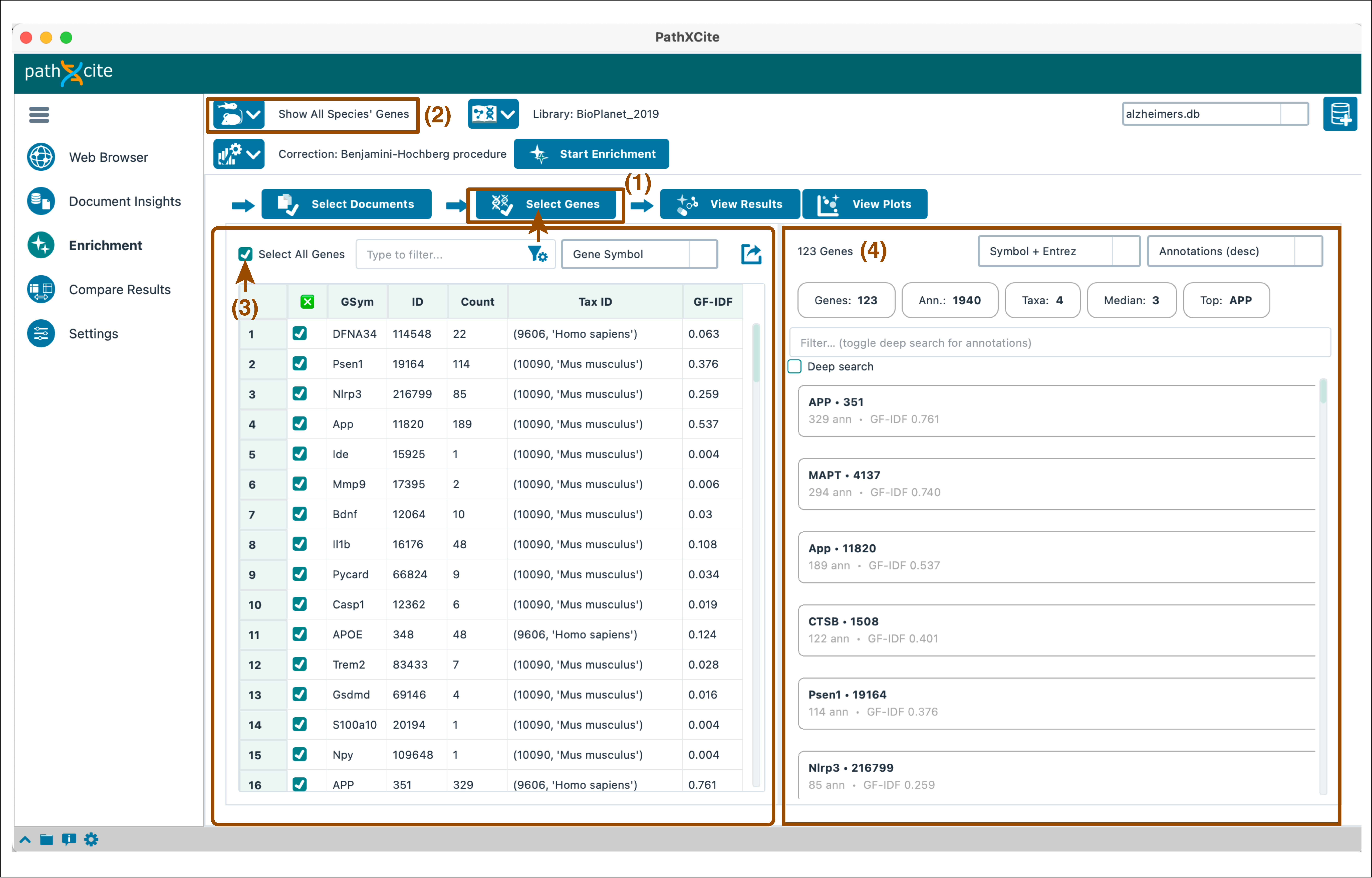
After selecting the documents, we can now choose the genes of interest.
- Click on Select Genes
- There is the option to filter the genes based on the respective species. For now, we will skip this step (See tutorial Gene Deep Dive for more details.)
- The table shows all genes mentioned in our selected articles, along with their Gene Symbol (GSym), Entrez ID (ID), Count (Absolute Number of Mentions), Taxonomy ID (Tax ID), as well as their GF-IDF Score (See tutorial Ranking Genes for more details.), For now, we will select all genes.
- Similar to the document selection, whenever you change the gene selection, the right view automatically updates. With a click on a gene (and then Show Sections), you can inspect in which textual contexts it is mentioned.
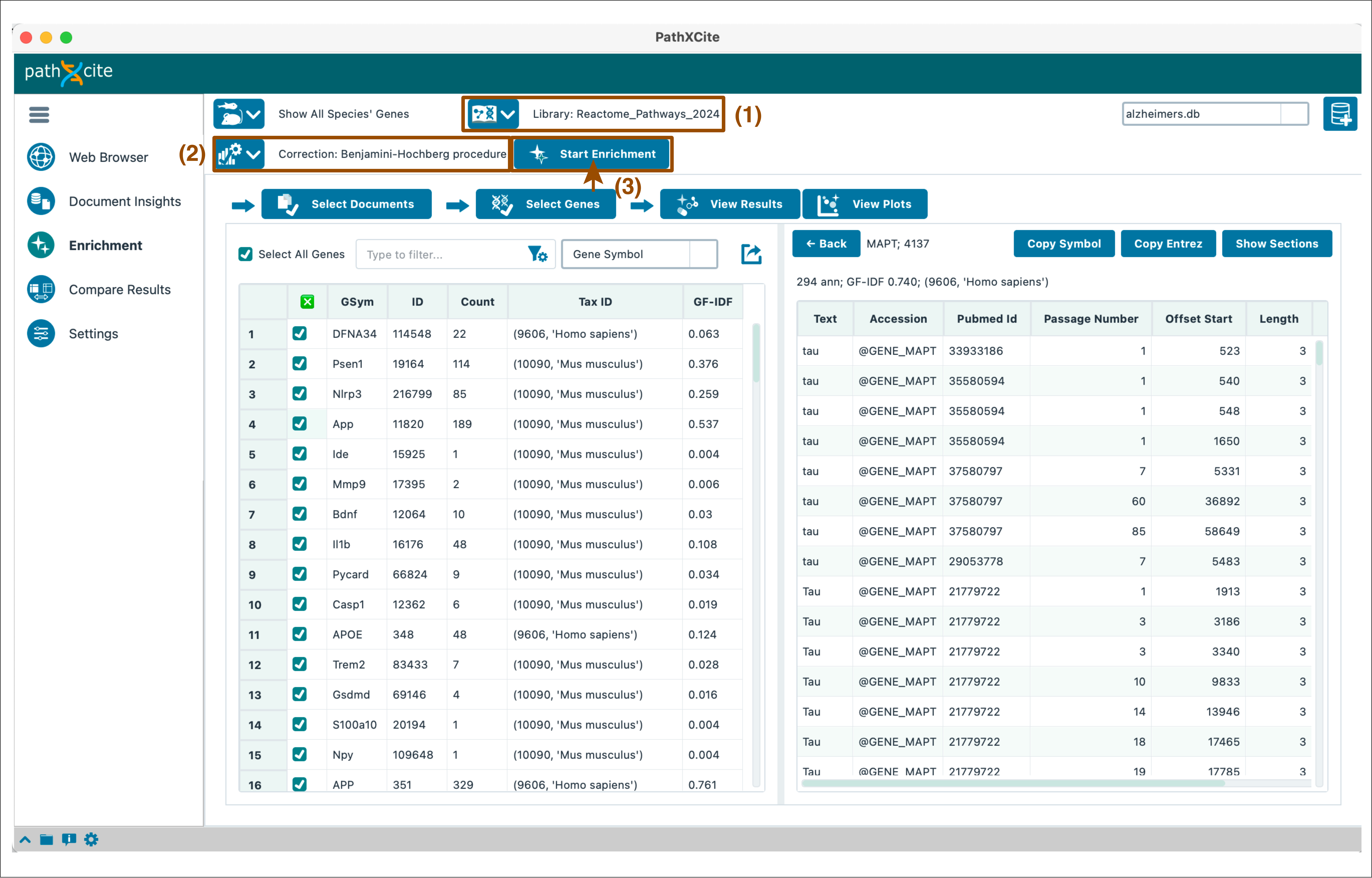
After selecting the documents and genes, we can now run the enrichment analysis.
- You can freely choose the gene set library against which you want to run the enrichment analysis. We selected Reactome Pathways 2024. But you can later on easily install and use over 240 predefined or your custom libraries (See tutorials Library manager, Custom Libraries, and Enrichr libraries for more details.).
- Also, you can adjust which correction method is used. For now, we will keep the default (Benjamini-Hochberg procedure, a common choice for multiple testing correction, see tutorial ORA and Corrections for more details).
- Now, you can click on Start Enrichment and within few seconds, the results will be displayed.
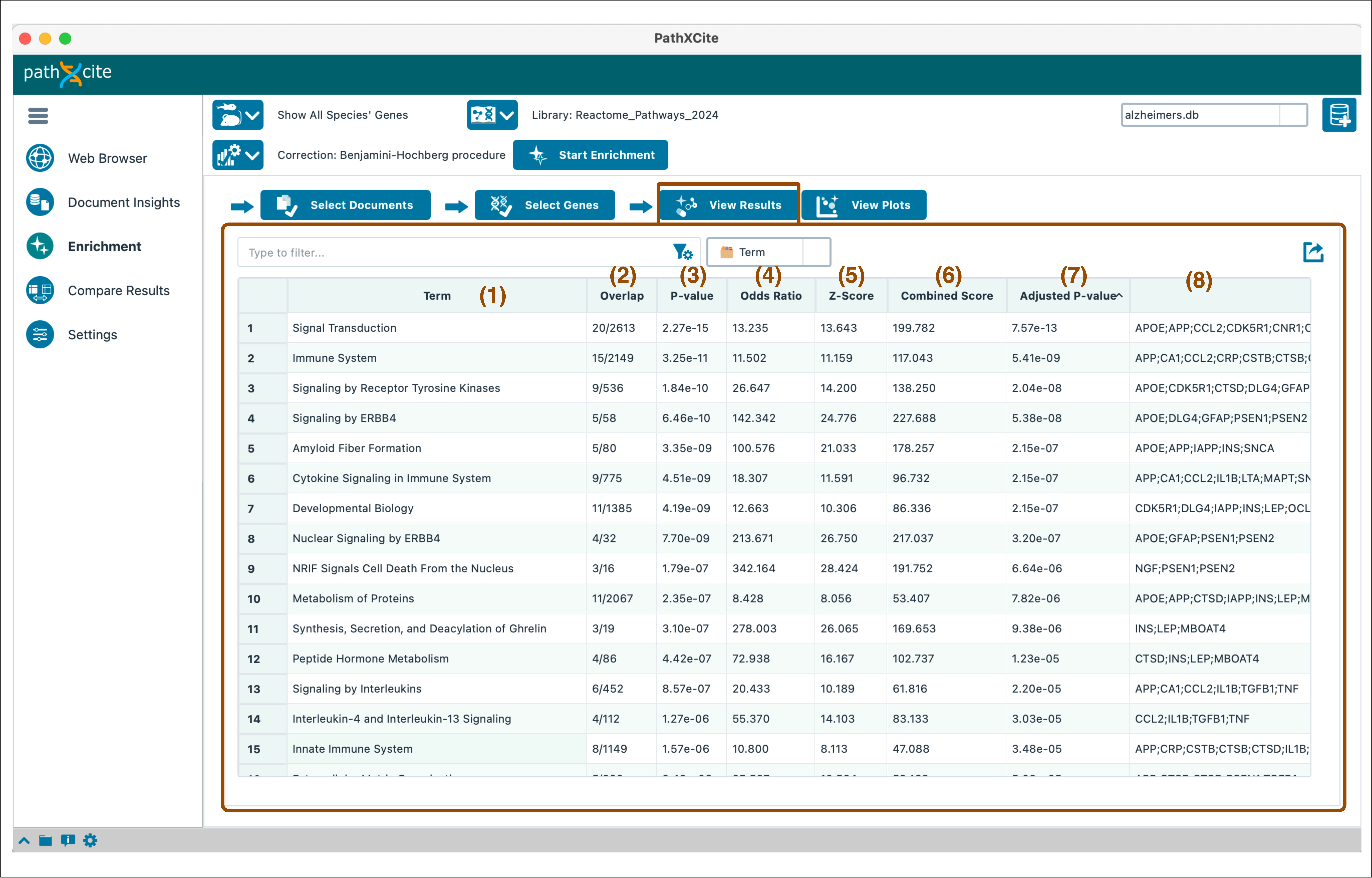
The enrichment analysis results are displayed in a table format.
- Terms: The biological terms or pathways that are enriched in your gene set (here, all Reactome pathways sharing at least one gene with our literature-derived genes).
- Overlap: The number of genes from your selected gene set that overlap with the genes in the term, along with the total number of genes in that term (e.g., 20/2613 means 20 out of 2613 genes in that pathway are present in your gene set).
- P-value: The statistical significance of the enrichment, indicating how likely it is to observe such an overlap by chance.
- Odds Ratio: The odds of observing the overlap versus not observing it.
- Z-score: A measure of how many standard deviations the observed overlap is from the expected overlap.
- Combined Score: A composite score that combines the p-value and z-score to provide a more comprehensive measure of enrichment significance.
- Adjusted P-value: The p-value adjusted for multiple testing using the selected correction method (here, Benjamini-Hochberg).
- Genes: The specific genes from your gene set that are part of the enriched term.
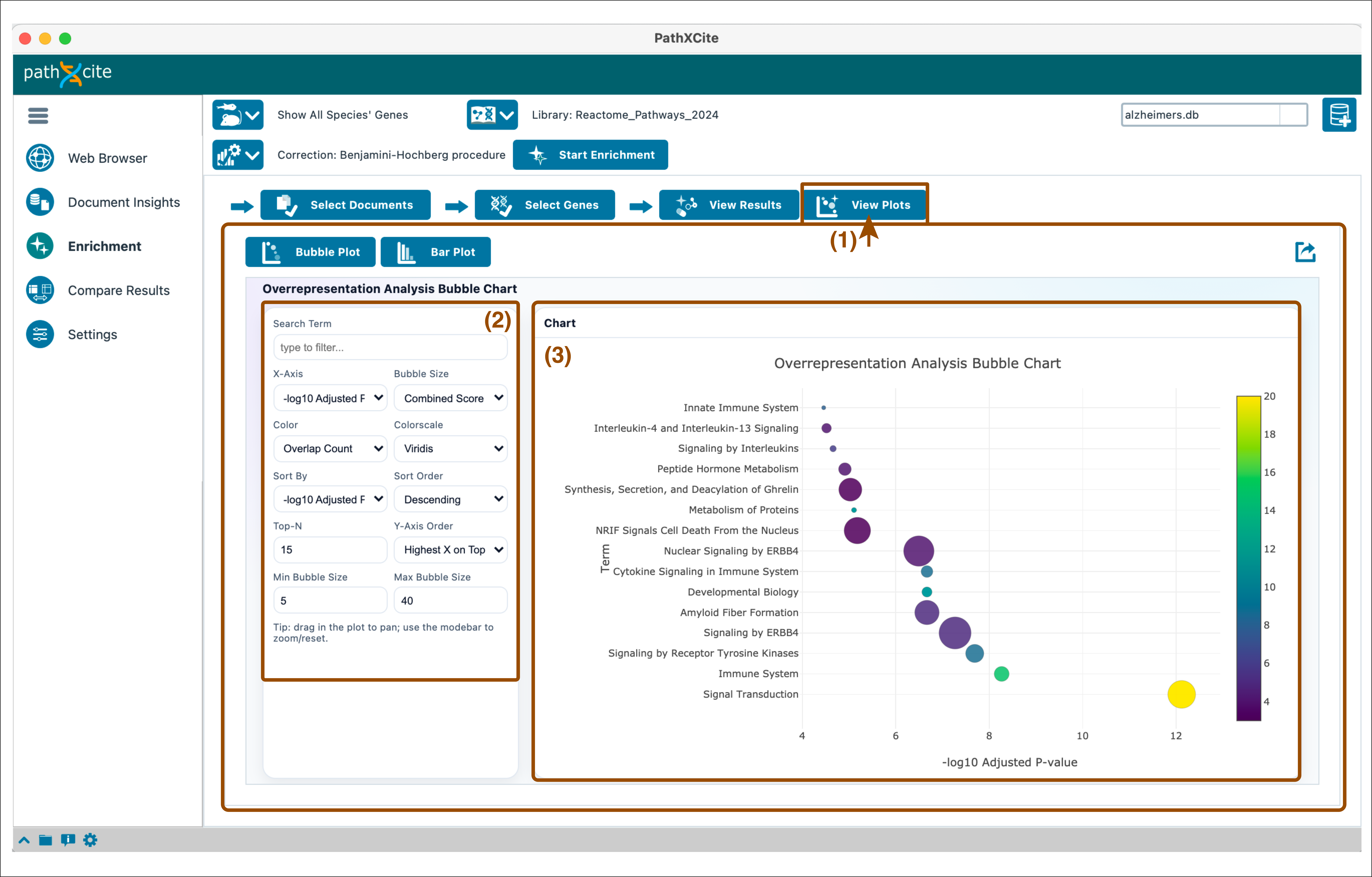
You can view the results either in an interactive bubble plot or bar plot.
- Click on View Plots to open the plot panel. It should first display the bubble plot.
- Use the controls to adjust the visualization settings, namely what the x-axis/bubble size/bubble color represents, how many terms are included, or the color scale is used.
- The plot is then directly updated.
