Step 1: Learn about Enrichr
Follow the steps below. Use ←/→ to navigate; press 1-9 to jump.
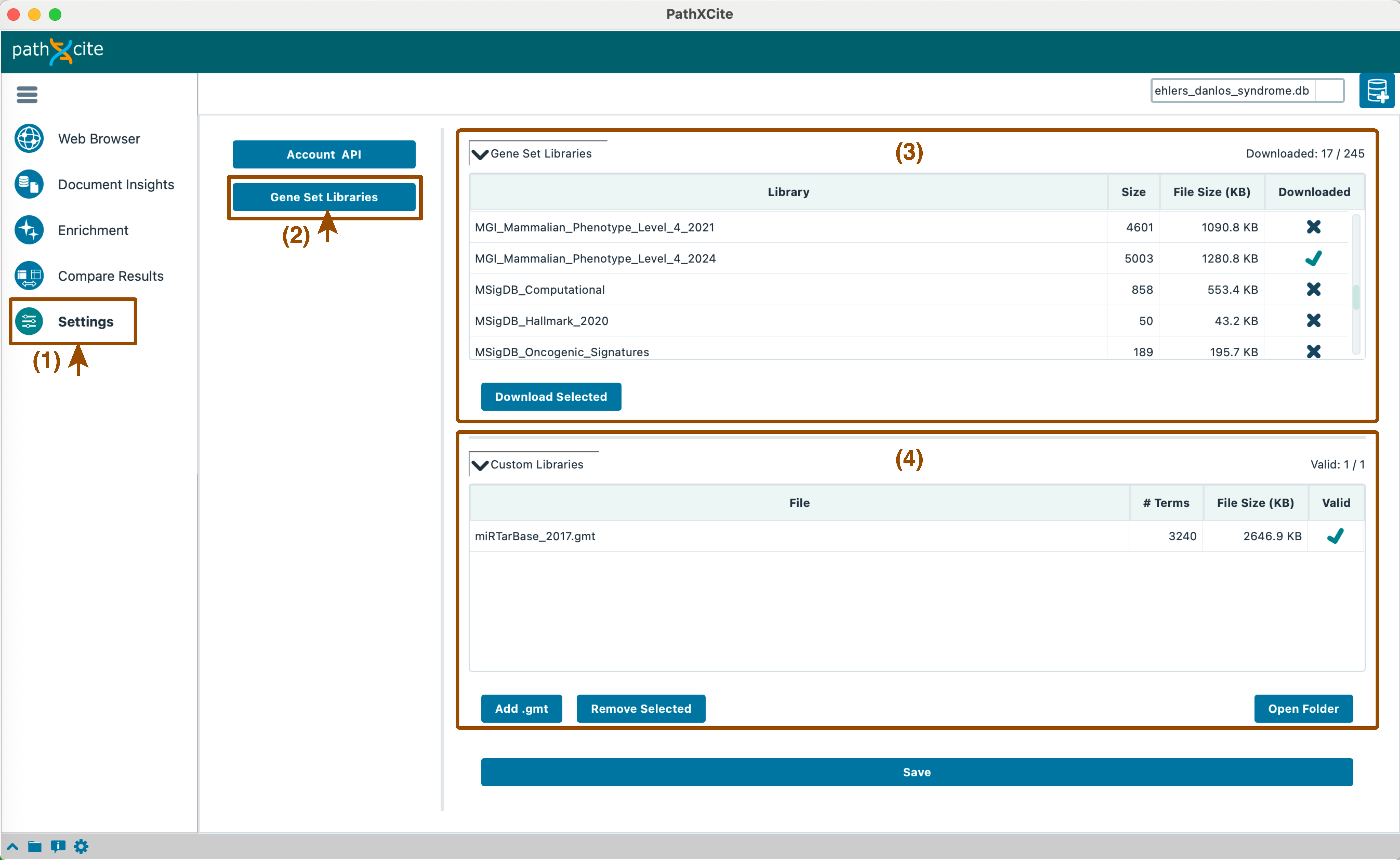
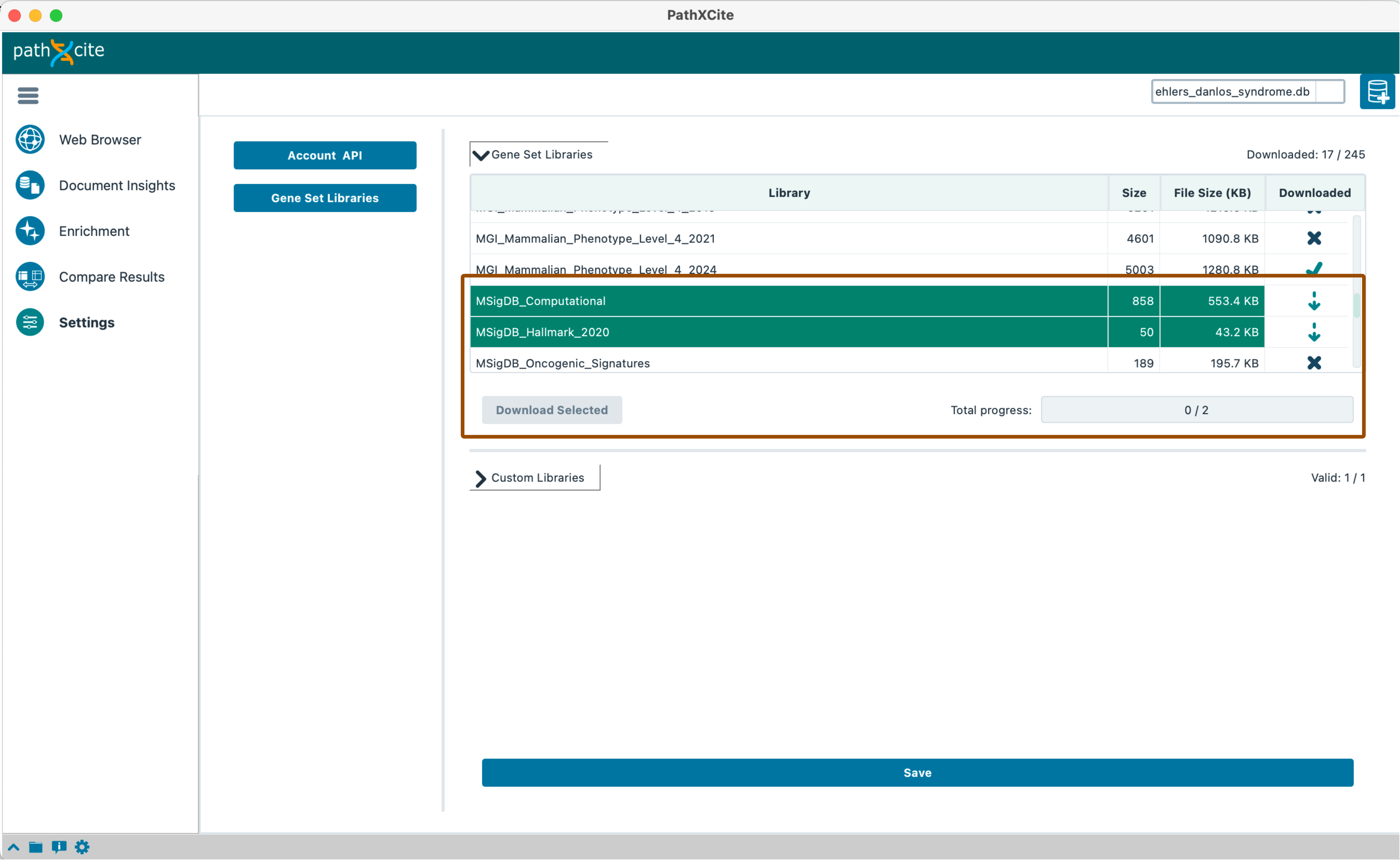
About Enrichr's predefined gene set libraries
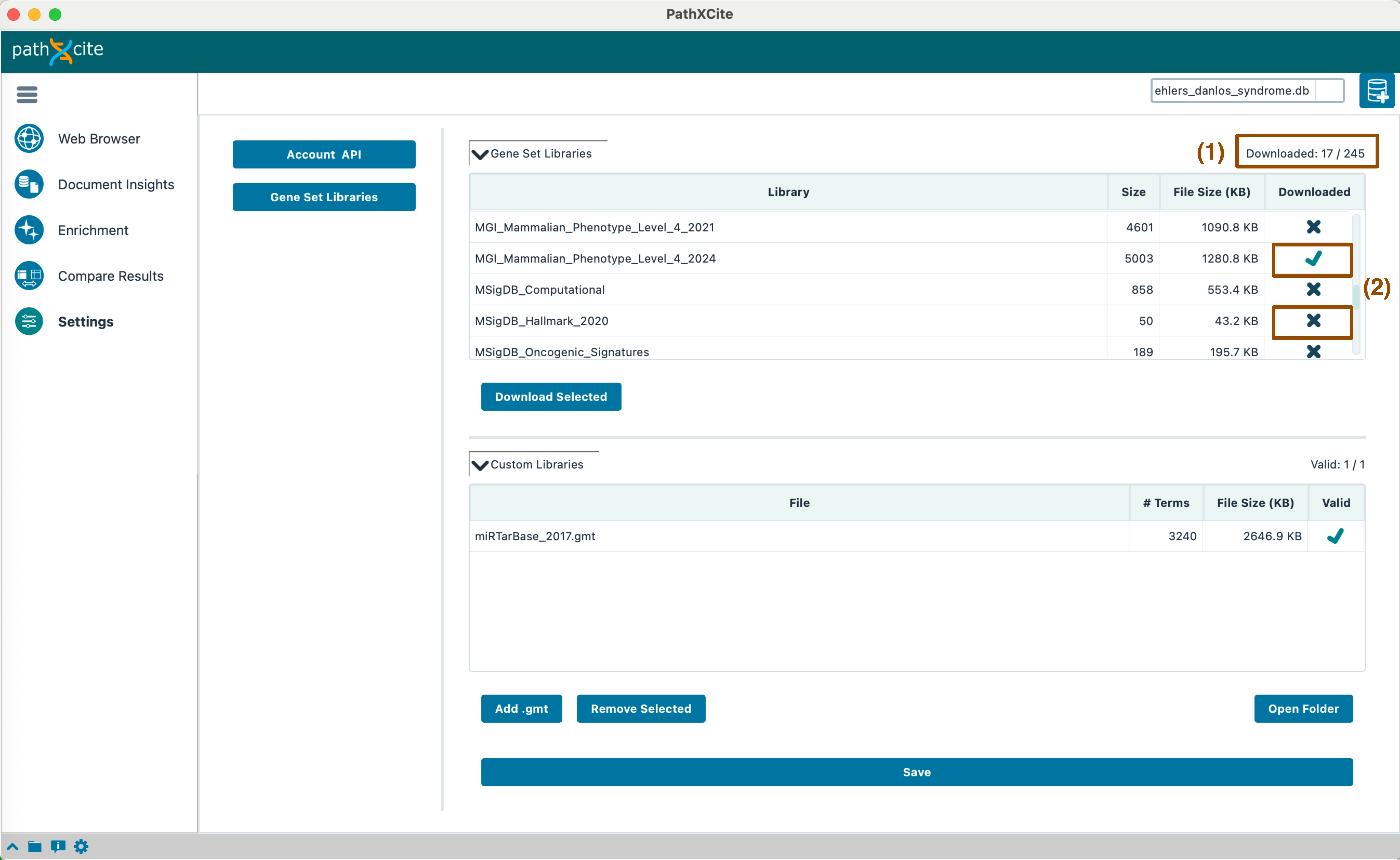
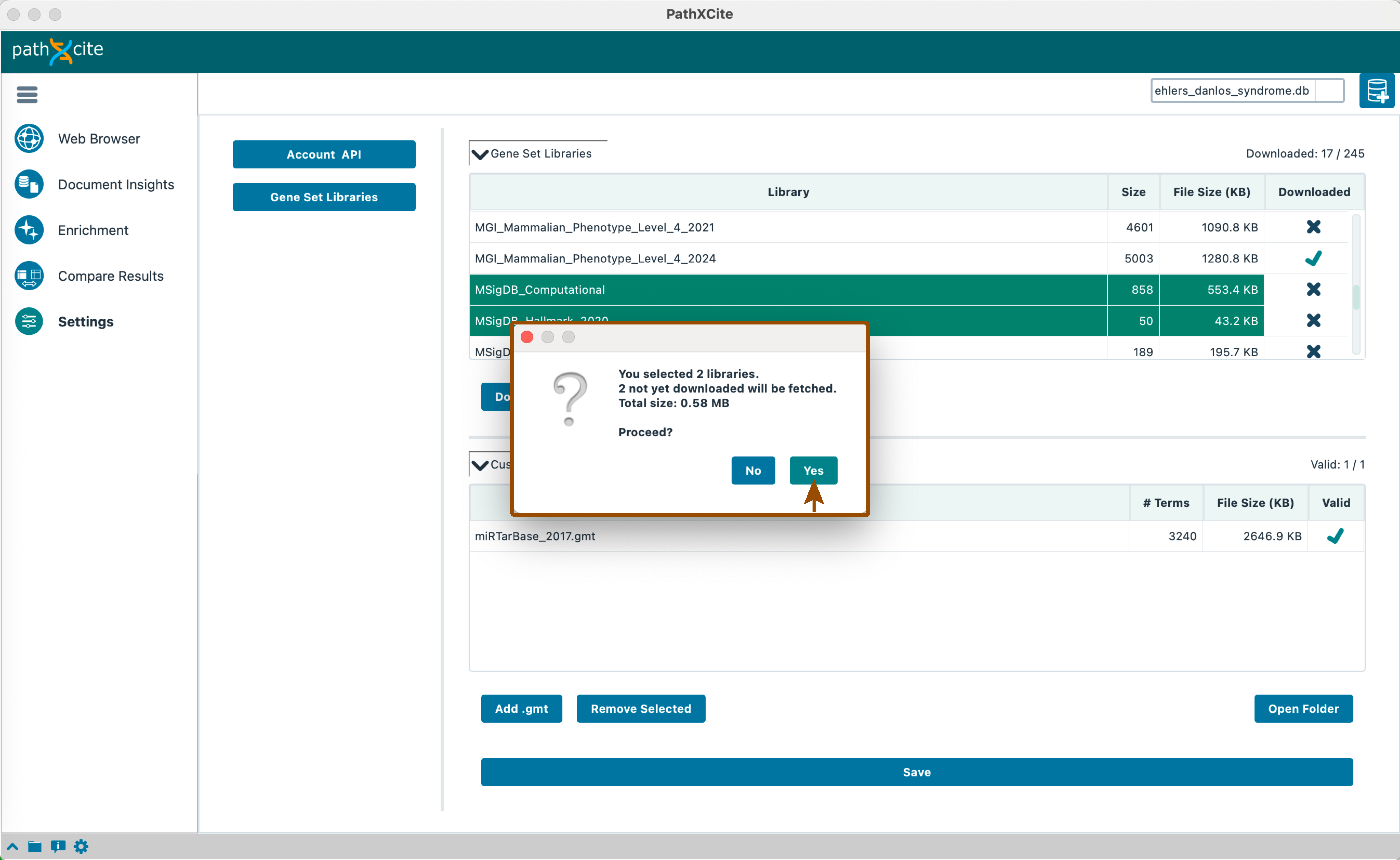
PathXcite lets you work with the predefined gene set libraries curated for Enrichr. It retrieves libraries on demand from Enrichr's public endpoint and stores them locally for fast, offline reuse.
Where libraries come from
Libraries are downloaded directly from Enrichr's API:
https://maayanlab.cloud/Enrichr/geneSetLibrary?mode=text&libraryName=<LIBRARY_NAME>libraryNameis any of Enrichr's predefined libraries (e.g.,GO_Biological_Process_2023,KEGG_2021_Human,ChEA_2022).mode=textreturns the library in tab-delimited GMT format (see below).- PathXcite caches the file locally after the first download so the same content is available across sessions.
Note: Enrichr maintains hundreds of libraries (≈245+ across categories). The exact catalog can evolve as resources are updated.
File format: GMT
Enrichr libraries are delivered as GMT (Gene Matrix Transposed) files, a compact, line-oriented format for gene set collections. Each line represents one gene set:
<SET_NAME> <GENE_1> <GENE_2> ... <GENE_N>- Fields are tab-separated.
- SET_NAME: a machine-friendly identifier (no tabs), for example a pathway name.
- GENE_i: a list of genes (here: HGNC symbols) that belong to the set.
Example (wrapped for display):
TNF_SIGNALING TNF TNFRSF1A TRADD TRAF2 MAP3K7 IKBKB NFKBIA RELA ...Common library types available in Enrichr
Enrichr aggregates many knowledge sources. Typical categories include:
- Gene Ontology (GO): Biological Process, Molecular Function, Cellular Component (multiple year-stamped releases).
- Pathways: KEGG, Reactome, WikiPathways, BioCarta.
- Transcription factor targets: ChEA, ENCODE, TRANSFAC-derived sets (chip-seq/TF motif-based target collections).
- Epigenomic signatures: ENCODE/ChEA/ROADMAP histone marks, open chromatin peaks.
- Protein complexes & interactions: CORUM, protein-protein interaction-derived modules.
- Disease & phenotype: DisGeNET, OMIM/Orphanet-inspired sets, HPO terms.
- Drug/perturbation signatures: DSigDB, LINCS L1000 / GEO up/down signatures, DrugMatrix-like resources.
- Cell types & tissues: cell atlas markers, tissue-enriched gene sets.
- Miscellaneous/other: miRNA targets, conserved motifs, aging, metabolism, and more.
The exact names and vintages (e.g., _2018, _2021, _2023) reflect the snapshot of each underlying database at the time of curation.
Local storage & versioning
- After download, libraries are saved as
.gmtfiles withinpathXcite/assets/external_data/gmt_files. - It's good practice to record the library name, download date, and (if available) source version for reproducibility.
- If analyses must be reproducible over time, pin library snapshots (e.g., keep the exact GMT used) instead of re-downloading latest.
Using the libraries in enrichment workflows
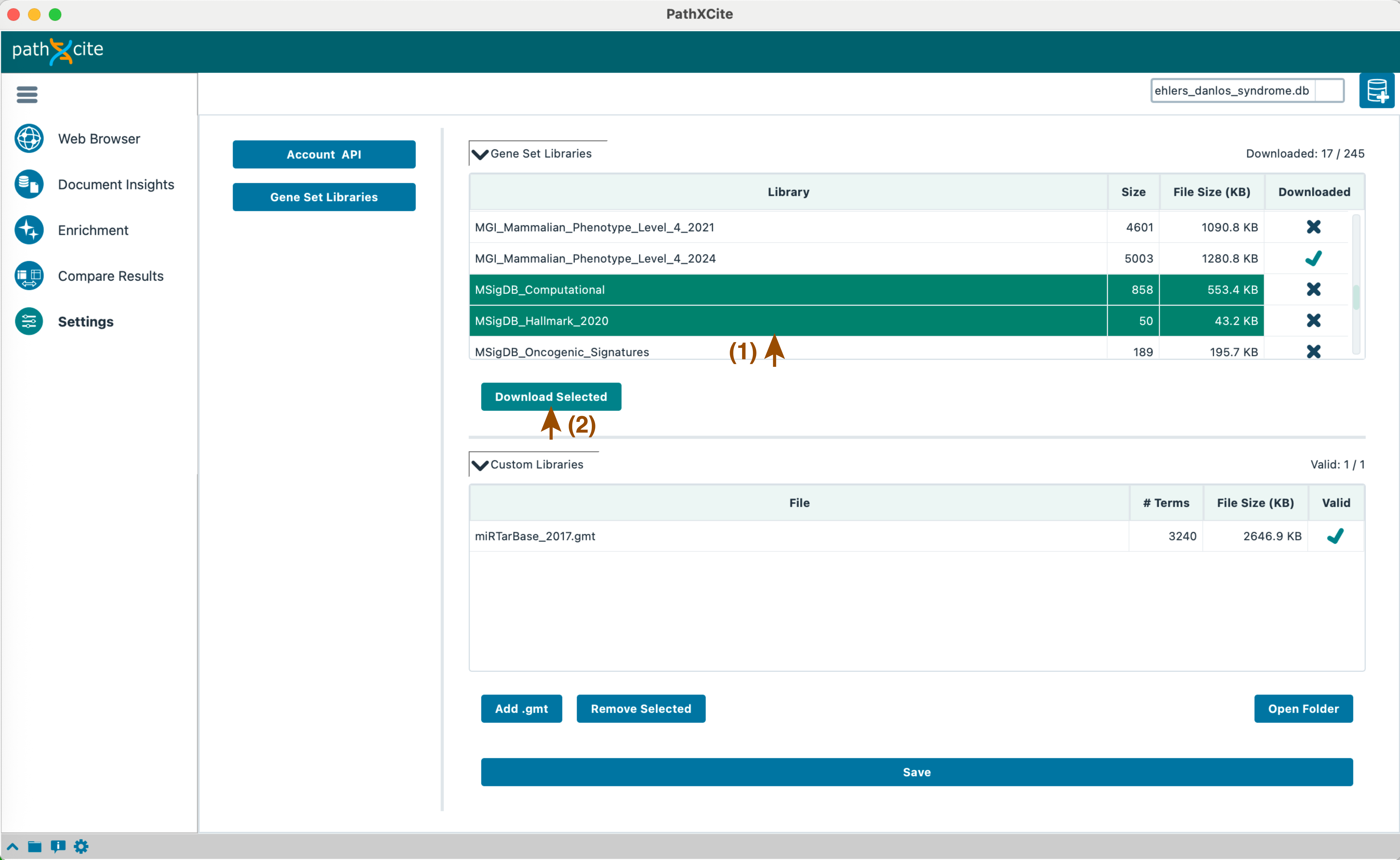
- Select a library whose biology matches your question (e.g., pathway libraries for mechanistic interpretation; TF-target libraries for regulatory hypotheses).
- Run enrichment using pathXcite.
- Adjust for multiple testing (e.g., Benjamini-Hochberg FDR) across all tested gene sets.
- Interpret results in biological context; corroborate with orthogonal evidence where possible.
Reference
Enrichr is maintained by the Ma'ayan Lab. Please cite as appropriate, for example:
- Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics (2013).
- Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res (2016).
- Xie Z, et al. Gene Set Knowledge Discovery with Enrichr. Curr Protoc (2021).
Website: maayanlab.cloud/Enrichr